Poji: a Fiji-based tool for analysis of podosomes and associated proteins
Podosomes are micron-sized cytoskeletal structures which enable the adhesion to and the degradation of extracellular matrix by a variety of cells, including macrophages. The structure of podosomes comprises a core of branched F-actin, a surrounding ring of integrins and adhesion plaque proteins (e.g. vinculin), a cap of regulators and nucleators on the top of the core, as well as acto-myosin fibers that both extend laterally along the core and between individual podosomes to connect them to superstructures. The size, amount and protein composition of podosomes can vary both in homeostasis (e.g. the size difference between successor and precursor podosomes) and in response to external cues like matrices of different stiffness or the treatment with inhibitors and chemical reagents. As macrophages regularly form >100 podosomes per cell, manual analysis of individual podosomes would be an insurmountable task. Thus, we developed a semi-automatic tool to quickly and reliably analyse individual podosomes, the combined podosome covered area as well as general properties of cells, both individually and in relation to each other. This tool is designed for the characterization of podosomes in fixed cell images (including z-stacks), but can, among others, also be used to detect invadopodia and to correlate them with the area of local matrix degradation.
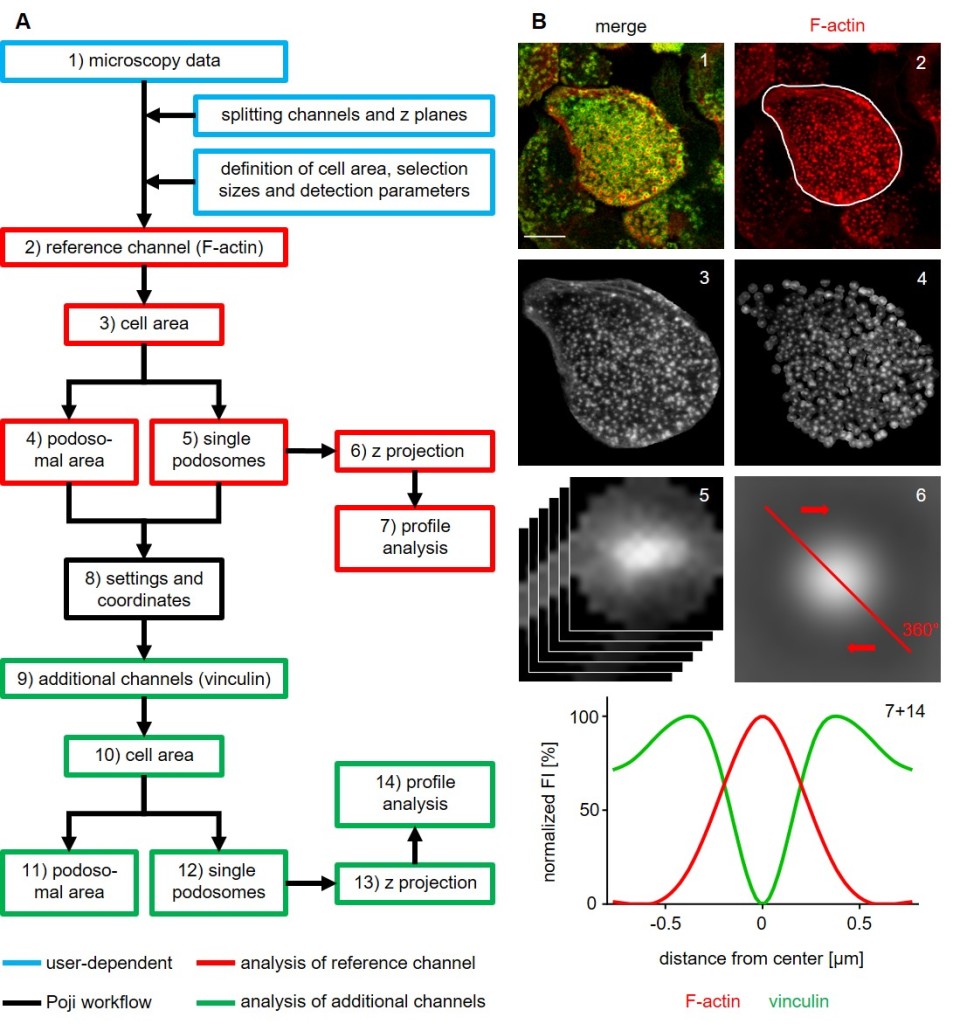
Schematic overview of the Poji work flow. (A) Main processing steps of the macro. The original image (1) is split and settings are defined prior to automated analysis (blue boxes; user-dependent steps). F-actin channel (2) is taken as a reference to determine cell parameters. Cell area (3), combined area of all podosomes (4) and individual podosomes (5) are analysed. Z projection of all podosomes (6) allows the creation of radial profile analyses (7). Selection coordinates (8) of all areas are saved and used in the analysis of additional fluorescence channels (9-14). (B) Corresponding images of the main processing steps. Primary human macrophage, stained for F-actin (podosome core) and for vinculin (podosome ring) (1). Successive steps of the analysis (2-6) and profile analysis of the z projection of podosomes (7+14). Bar: 10 µm. (Herzog et al. 2020, Journal of Cell Science).
While the analysis of cells is conducted in automated processes, the setting of predefined parameters is user-dependent. These parameters comprise the manual definition of the cell area (with an optional additional selection of podosome clusters) and settings for podosome detection and further processing. While the splitting of z slices and channels (automatable by using an additional, optional macro) has to be done prior to starting the macro, all other settings are defined in interfaces of the macro. The settings include defining the noise threshold for local maxima recognition, as well as smoothing of images, to better detect the geometrical centre of podosomes. For further advices on using the macro, please refer to the user guide that is deposited together with the codes and premade analysis tables (see bottom of the page).
Analysis of images by using Poji can yield up to 19 different sets of data, that describe properties of the entire cell and of podosomes (collectively and individually) as well as calculating ratios between them. Examples of these calculated results are shown below.

Examples for analysis results. (A,B) Human primary macrophage, stained for F-actin (red), myosin IIA (grey) and vinculin (green) as control (A) or after treatment with cytochalasin D (B). (C,D) General cell parameters like cell size (C) and number of podosomes per cell (D). (E,F) Fluorescence intensity measurements showing the prevalence of the 3 protein stainings in the cell area (E) and per podosome (F). (G,H) Profile analysis of the mean of all Podosomes of control (G) and treated cells (H). Results from n≥15 cells with mean ± S.E.M. Click image to enlarge.
This tool was originally published in:
Herzog et al., „Poji: a Fiji-based tool for analysis of podosomes and associated proteins“, Journal of Cell Science, 2020.
The code, together with associated premade analysis tables and an extensive user guide, is available online at:
https://github.com/roherzog/Poji
